Principal Component Analysis (PCA) is a statistical technique used to reduce the dimensionality of a data set while retaining as much of the variation in the data as possible. It does this by finding a new set of uncorrelated variables, called principal components, which can be used to represent the original data.
To perform PCA in R, the first step is to load the data and scale it if necessary. Next, the prcomp() function can be used to perform the analysis, which returns an object containing the principal component scores and loadings. The summary() function can be used to view the results, and the biplot() function can be used to create a graphical representation of the data in the new principal component space.
Here is an example of performing PCA in R:
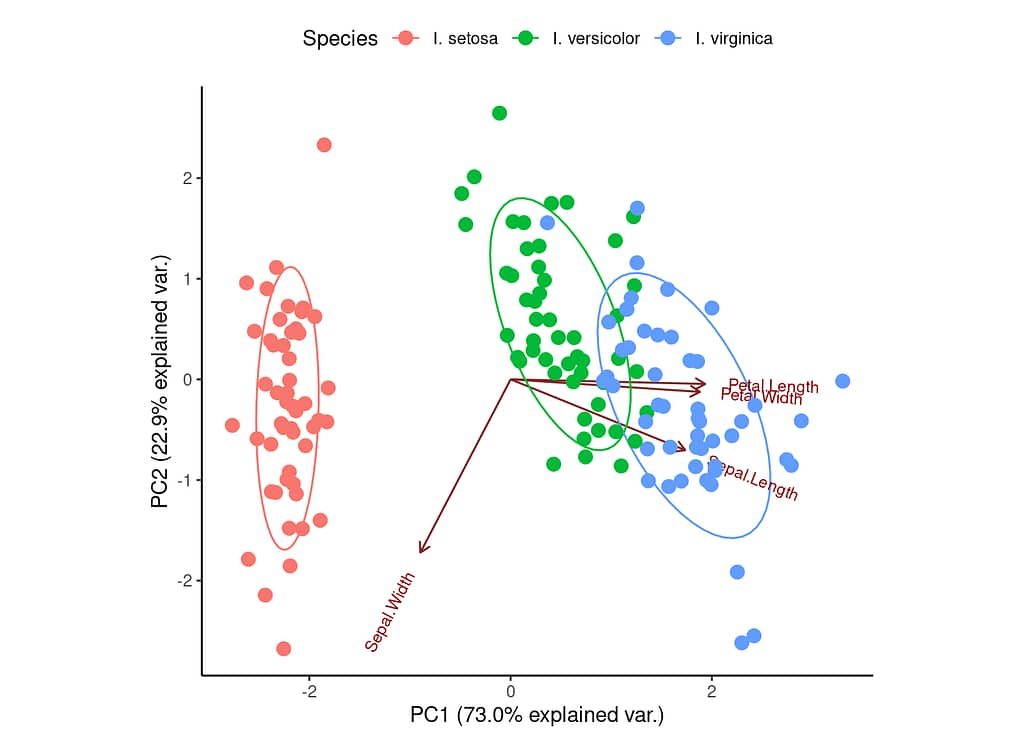
You can see that the first principal component (PC1) explains 73% of the variance in the data. The next principal component (PC2) explains an additional 22.9%. The ellipses represent potential groupings of data based on the principal components. In the example above, we can see that the Iris species form three relatively clear clusters (particularly I. setosa).
We can do something similar in Python:
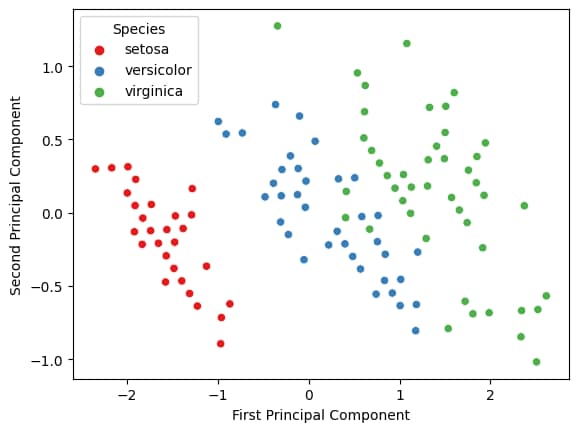
You might notice that there is a lot more information provided in the R plot, but more importantly, the results are the same for both PCAs, regardless of which programming language we use.